Unique structural features of microRNAs
In the last two decades, the small non-coding RNA field has significantly expanded beyond snRNAs & snoRNAs (1) to include piRNAs (2–4), siRNAs (5, 6), novel small RNAs derived from known non-coding RNAs including tRNAs (7) and, of course, microRNAs (8–11). Each of these types of small RNAs are characterized by a distinctive suite of characteristics and a unique evolutionary history. microRNAs can be distinguished from other small genomically encoded RNA families by the following criteria (12–14) & Figure):
Two 20-26nt long reads are expressed from each of the two arms derived from a hairpin precursor.
Because the ends of canonical microRNA reads are generated enzymatically, the 5’ ends of the reads are homogeneous (>90%).
The hairpin precursor shows imperfect complementarity, and base pairs in at least 16 of the ~ 22 nucleotides.
The 5p and 3p reads are offset by 2 nucleotides on both ends due to the sequential processing of the microRNA transcript by Drosha and Dicer to generate the mature ~22 nucleotide read(s). In some cases, the Drosha offset is only offset by 1 templated nucleotide, but in these cases the 3’ end of the 3p arm is monouridilyated (15, 16).
The length of the loop is at least 8 nucleotides long; there is no apparent maximum in loop length, even in organisms possessing only a single Dicer gene, contra (13), even though most taxa like vertebrates with single Dicer genes never show loop lengths greater than ~40 nucleotides.
The mature microRNA sequence usually starts with A or U, and is often mismatched with the complementary arm, which seems to facilitate arm selection by Argonaute (at least in mammals) (17,18,19).
Nucleotide positions 2 through 8 of the mature sequence (the "seed") are strongly conserved through evolution, as are positions 13-16 (the 3' complementary region) (13,20,21).
Processing motifs are often (but not always) present in the primary microRNA transcript including a UG motif 14 nucleotides upstream of the 5p arm, a UGU motif at the 3' end of the 5p arm, and a CNNC motif 17 nucleotides downstream of the 3p arm (22,23).
There are other features of microRNAs, in particular structural and evolutionary signatures, that allow them to be further distinguished from other small RNAs:
Recognition and utilization of clear and, for the most part mechanistically well understood, criteria will allow the delineation of bona fide microRNAs from the myriad small RNAs generated in eukaryotic cells, allowing for deeper and robust insights into their function, possible mis-regulation, and evolution.
References
1. Matera,A.G., Terns,R.M. and Terns,M.P. (2007) Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol., 8, 209–220.
2. Girard,A., Sachidanandam,R., Hannon,G.J. and Carmell,M.A. (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature, 442, 199–202.
3. Aravin,A., Gaidatzis,D., Pfeffer,S., Lagos-Quintana,M., Landgraf,P., Iovino,N., Morris,P., Brownstein,M.J., Kuramochi-Miyagawa,S., Nakano,T., et al. (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature, 442, 203–207.
4. Grivna,S.T., Beyret,E., Wang,Z. and Lin,H. (2006) A novel class of small RNAs in mouse spermatogenic cells. Genes Dev., 20, 1709–1714.
5. Caplen,N.J., Parrish,S., Imani,F., Fire,A. and Morgan,R.A. (2001) Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. U. S. A., 98, 9742–9747.
6. Grishok,A., Pasquinelli,A.E., Conte,D., Li,N., Parrish,S., Ha,I., Baillie,D.L., Fire,A., Ruvkun,G. and Mello,C.C. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–34.
7. Lee,Y.S., Shibata,Y., Malhotra,A. and Dutta,A. (2009) A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev., 23, 2639–2649.
8. Pasquinelli,A.E., Reinhart,B.J., Slack,F., Martindale,M.Q., Kuroda,M.I., Maller,B., Hayward,D.C., Ball,E.E., Degnan,B., Müller,P., et al. (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature, 408, 86–89.
9. Lagos-Quintana,M., Rauhut,R., Lendeckel,W. and Tuschl,T. (2001) Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858.
10. Lau,N.C., Lim,L.P., Weinstein,E.G. and Bartel,D.P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 294, 858–862.
11. Lee,R.C. and Ambros,V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science, 294, 862–864.
12. Fang,W. and Bartel,D.P. (2015) The Menu of Features that Define Primary MicroRNAs and Enable De Novo Design of MicroRNA Genes. Mol. Cell, 60, 131–145.
13. Fromm,B., Billipp,T., Peck,L.E., Johansen,M., Tarver,J.E., King,B.L., Newcomb,J.M., Sempere,L.F., Flatmark,K., Hovig,E., et al. (2015) A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu. Rev. Genet., 49, 213–242.
14. Bartel,D.P. (2018) Metazoan MicroRNAs. Cell, 173, 20–51.
15. Kim,B., Ha,M., Loeff,L., Chang,H., Simanshu,D.K., Li,S., Fareh,M., Patel,D.J., Joo,C. and Kim,V.N. (2015) TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J., 34, 1801–1815.
16. Kim,Y.-K., Kim,B. and Narry Kim,V. (2016) Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. U. S. A., 113, E1881–E1889.
17. Schwarz,D.S., Hutvágner,G., Du,T., Xu,Z., Aronin,N. and Zamore,P.D. (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell, 115, 199–208.
18. Khvorova,A., Reynolds,A. and Jayasena,S.D. (2003) Functional siRNAs and microRNAs exhibit strand bias. Cell, 115, 209–216.
19. Suzuki,H.I., Katsura,A., Yasuda,T., Ueno,T., Mano,H., Sugimoto,K. and Miyazono,K. (2015) Small-RNA asymmetry is directly driven by mammalian Argonautes. Nat. Struct. Mol. Biol., 22, 512–521.
20. Grimson,A., Farh,K.K.-H., Johnston,W.K., Garrett-Engele,P., Lim,L.P. and Bartel,D.P. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell, 27, 91–105.
21. Wheeler,B.M., Heimberg,A.M., Moy,V.N., Sperling,E.A., Holstein,T.W., Heber,S. and Peterson,K.J. (2009) The deep evolution of metazoan microRNAs. Evol. Dev., 11, 50–68.
22. Auyeung,V.C., Ulitsky,I., McGeary,S.E. and Bartel,D.P. (2013) Beyond secondary structure: primary-sequence determinants license pri-microRNA hairpins for processing. Cell, 152, 844–858.
23. Nguyen,T.A., Jo,M.H., Choi,Y.G., Park,J., Kwon,S.C., Hohng,S., Kim,V.N. and Woo,J.S. (2015) Functional Anatomy of the Human Microprocessor. Cell, 161, 1374–1387.
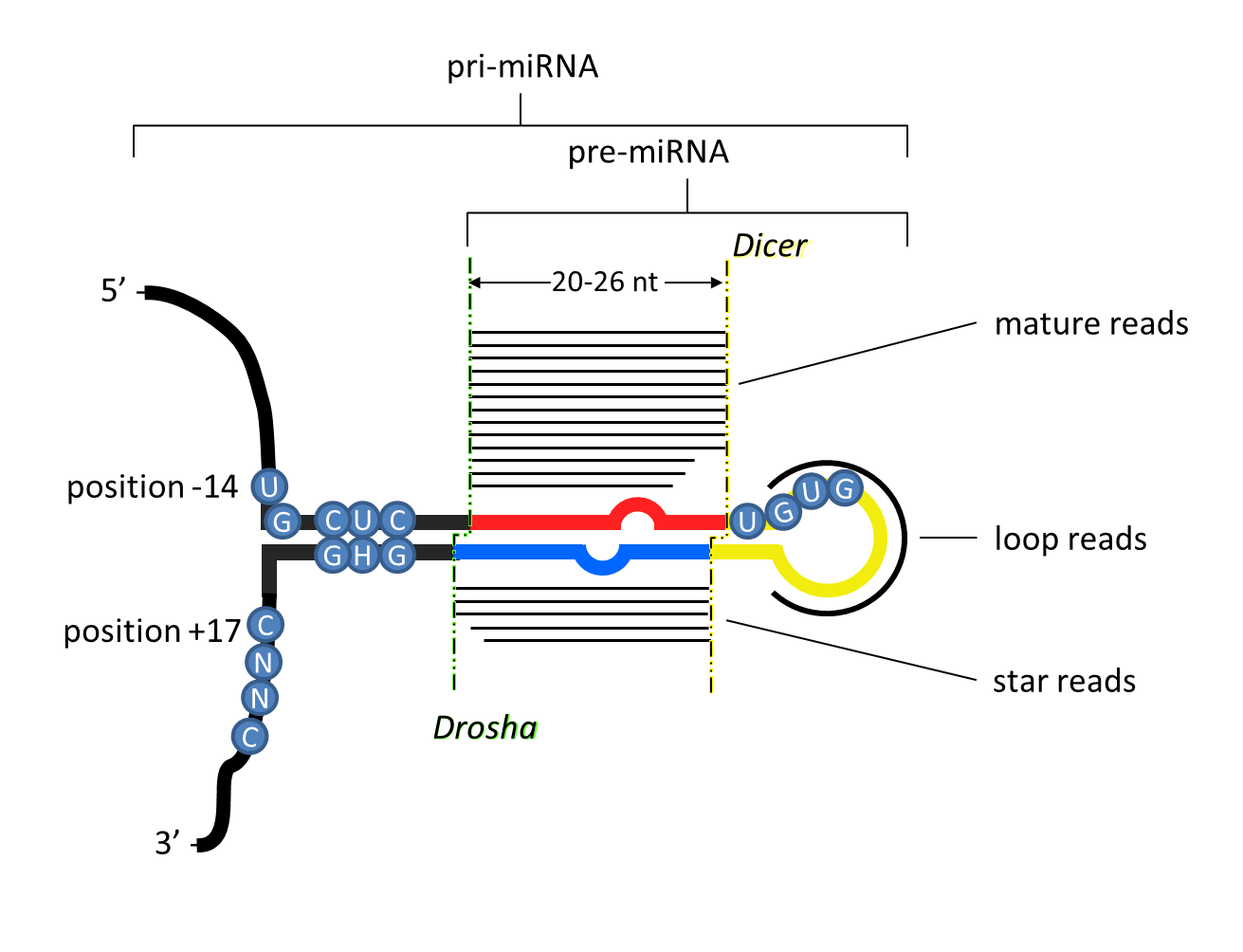
Figure: unique criteria of microRNAs
Contact:






